Automated Data Preprocessing
Imputation of missing values and batch effect correction to make the expression matrix statistically significant
A platform bridging machine learning and omics-biology to unveil the encrypted language of life.

Joy Prokash Debnath, Kabir Hossen, Md. Sayeam Khandaker, Shawon Majid, Md Mehrajul Islam, Siam Arefin, Preonath Chondrow Dev, Saifuddin Sarker, Tanvir Hossain
bioRxiv, Cold Spring Harbor Laboratory (2025)
DOI: 10.1101/2025.10.25.684517
View PDF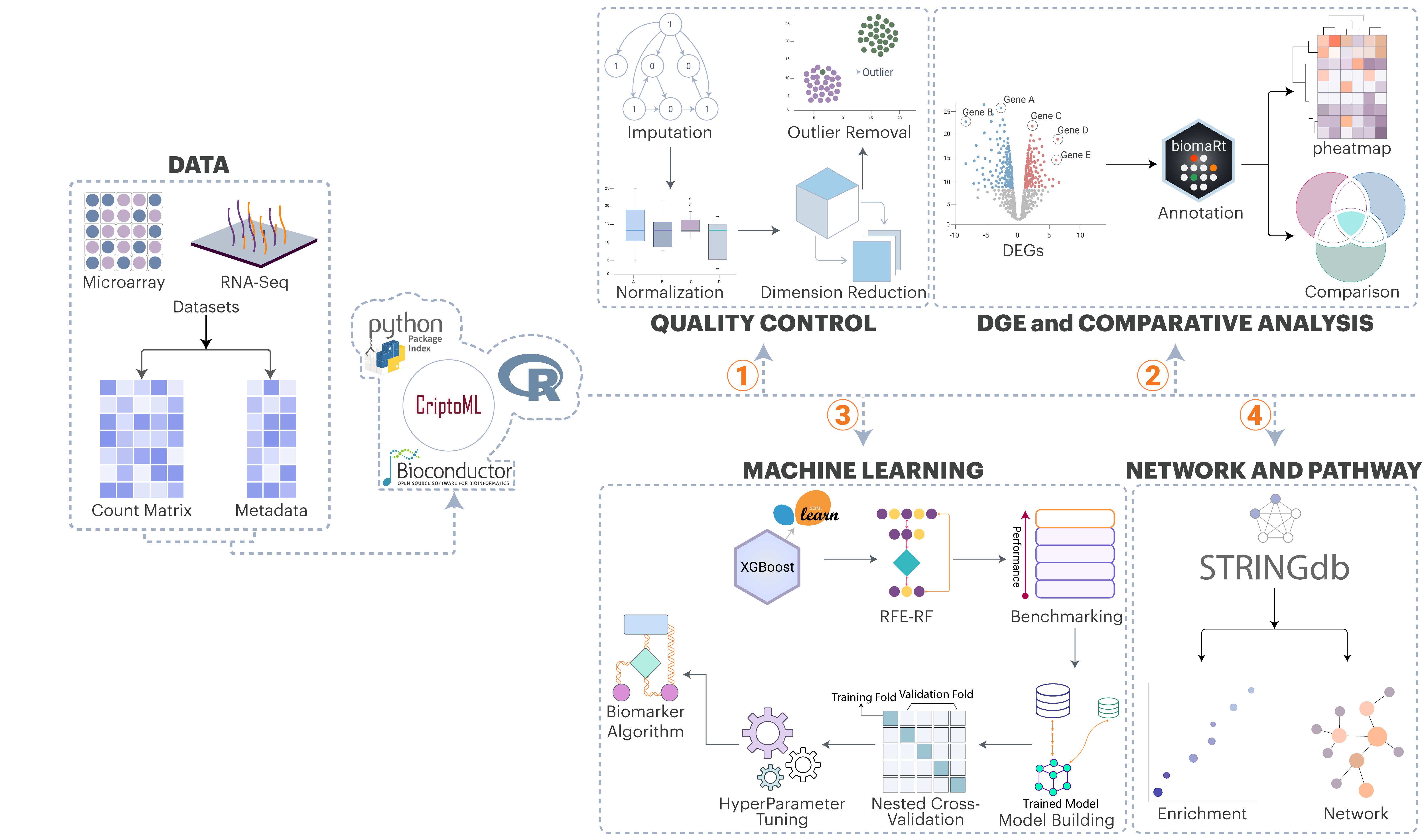
Transcriptomic biomarker discovery has been a challenge due to variation in datasets and platforms, complexity in statistical and computational methods, integration of multiple programing languages, intricacy of ML workflow to evaluate biomarkers. Standard workflows necessitate several stages (quality control, normalization, differential expression), typically executed in R or Python, resulting in bottlenecks for non-experts.
We present omicML, an intuitive graphical user interface (GUI) that combines transcriptomic data analysis with machine learning (ML)-based classification via integrating R and Python packages/libraries. It supports both RNA-Seq and microarray data, automating preprocessing and differential expression analysis. Our extensive ML pipeline enables both supervised and unsupervised learning, integrates various datasets based on candidate gene signatures, and systematically finalizes the biomarker algorithm.
In a case study, omicML identified a six-gene diagnostic model that distinguishes Mpox (monkeypox virus) infections from those caused by other viruses, including SARS-CoV-2, HIV, Ebola, and varicella-zoster. These results illustrate omicML's capacity to discern clinically relevant biomarkers from complex transcriptome data.
Integrating data normalization, differential gene expression analysis, annotation, heatmap analysis, dataset integration, batch effect removal, machine learning analysis, and functional analysis into a unified system diminishes technical barriers and accelerates the conversion of expression data into diagnostic insights for clinicians and bench scientists.
Core capabilities of the OmicML framework
Imputation of missing values and batch effect correction to make the expression matrix statistically significant
both RNA-Seq and microarray datasets, enabling cross-platform analysis
Broad taxonomic coverage for gene level annotation across 367 species
Data standardization, feature selection, benchmarking, nested cross-validation, hyperparameter tuning, feature importance, single-gene model building, multi-gene model (biomarker algorithm) building
Gene-model based identification of biomarker for distinct conditions
Contextualization of the candidate biomarkers within biological pathways

The researchers and developers behind OmicML









Note: Package versions are periodically updated to ensure compatibility and access to the latest features.
To address the gaps in OmicML v1.0, future development will prioritize the introduction of additional modules and data types. Proposed improvements encompass gene co-expression networks, survival analysis (notably for cancer cohorts), deep-learning frameworks, integration of proteomics data, single-cell RNA-Seq analysis, ChIP-Seq data processing, spatial transcriptomics, multi-omics integration, AI-driven multi-omics modelling, single-cell ATAC-Seq analysis, specialized modules for bulk RNA-Seq, pan-cancer comparative analyses, and AI applications in genomics, primer design, and PCR data analysis. This enhanced capability will establish omicML as a multifaceted and robust instrument for translational omics analysis.